Cas9 Protein with NLS
Cas9 Protein with Dual NLS – Rapid, Precise Genome Editing with RNP Delivery
Accelerate your genome editing workflows with high-purity Cas9 protein featuring dual nuclear localization signals (NLS) for efficient nuclear import. Simply incubate the Cas9 protein with your guide RNA (gRNA or sgRNA) to form an active Cas9 ribonucleoprotein (RNP) complex—ready for immediate delivery into cells or embryos.
⚡Why Choose Cas9 Protein Over Plasmid or mRNA Delivery?
- Immediate Editing Activity: Cas9 protein starts working as soon as it enters the cell—no transcription or translation delay.
- Reduced Off-Target Effects: Degraded within ~24 hours, minimizing prolonged exposure and reducing the risk of mosaicism.
- No Genomic Integration: Unlike plasmid-based delivery, Cas9 protein avoids random DNA integration and persistent expression.
🌎Broad Compatibility
Because sgRNA/Cas9 RNP delivery bypasses host transcription and translation, it is effective in a wide range of species:
- Human, mouse, rat
- Zebrafish, nematodes, frogs
- Insects such as fruit flies, butterflies, mosquitoes
- Plants and other non-model organisms
✅Key Features
- Dual NLS for enhanced nuclear import
- Ready-to-use: Ideal for direct microinjection or transfection
- Validated across a range of in vivo and in vitro applications
- Endotoxin-free, high purity (>90%)
- Strict quality control for lot-to-lot consistency
- Available in flexible pack sizes
- Cost-effective without compromising quality
📬Need Help or Have Questions?
Our technical team is happy to assist.
📧 Contact us at info@pnabio.com.
“We guarantee the highest activity and the lowest pricing on Cas9 proteins.”
PNA Bio’s Cas9 protein is derived from Streptococcus
Our Cas9 protein is shipped the day of order by 5 pm ET for the next day delivery.
WT Cas9 with dual NLS, Standard Concentration – CP01
- Lyophilized Cas9 protein
- Sizes: 50 µg, or 200 µg (4 × 50 µg)
- Reconstitutes to 1 mg/mL, 6 μM if dissolved in 50 μL nuclease free water
- Recommended for microinjection and in vitro cleavage assay
WT Cas9 with dual NLS, High Concentration – CP02 & CP03
- Lyophilized Cas9 protein
- CP02: 250 µg
- CP03: 1 mg (4 × 250 µg)
- Reconstitutes to 5 mg/mL, 30 μM if dissolved in 50 μLnuclease free water
- Recommended for electroporation and in vitro cleavage assay
WT Cas9 with dual NLS, Liquid Form – CP04 Series
- CP04-100: 1 mg/mL, 20 µg × 5
- CP04-500: 5 mg/mL, 100 µg × 5
- Shipped same day for next-day delivery
Buffers & Reagents
- CB01:10× Cas9 reaction buffer for in vitro cleavage assays (500 µL)
- CB03:Cas9 dilution buffer for adjusting protein concentration (1 mL)
Nickase Cas9 (D10A mutant) – CN01 & CN02
- Lyophilized form
- CN01: 50 ug, reconstitutes to 1 mg/mL if dissolved in 50 μL water
- CN02: 5 mg/mL, 250 µg, reconstitutes to 5 mg/mL if dissolved in 50 μL water
- Ideal for generating nicks in DNA rather than double-strand breaks
- Can be beneficial for generating KI mutations
Dead Cas9 (dCas9, D10A/H840A mutant) – CD01 & CD02
- CD01: 1 mg/mL, 50 µg
- CD02: 1 mg/mL, 50 µg x 5 tubes
- Reconstitutes to 1 mg/mL if dissolved in 50 μL water
- Use for transcriptional repression or imaging applications
SniperCas9 – High-Fidelity Cas9 Variant
- Designed via directed evolution for improved specificity
- Lyophilized form
- CS01: 1 mg/mL, 50 µg or 200 µg (50 µg x 4 tubes)
- CS02: 5 mg/mL, 250 µg
- Reference: Directed evolution of CRISPR-Cas9 to increase its specificity. Lee JK et al. (2018) Nature Comm 9:3048.
Cy3-Labeled WT Cas9 Protein – CP06
- WT SpCas9 with NLS, conjugated to a Cy3 red fluorophore
- Enables fluorescence-based sorting and visualization of transfected cells
- Activity validated through in vitro cleavage assays and cell transfection
- Available in 50 µg, 100 µg, or 250 µg sizes
- Supplied at a concentration of 5 mg/mL
Available Buffers
– 10× Cas9 Reaction Buffer – For in vitro cleavage assays to evaluate guide RNA efficiency
– Cas9 Dilution Buffer – To adjust Cas9 protein to lower working concentrations
| Product | Cat No | Size | Format | After reconstitution |
|---|---|---|---|---|
| Standard Conc WT Cas9 (1 mg/ml after reconstitution) |
CP01-50 | 50 ug (300 pmol) | Lyophilized | 6 uM x 50 ul |
| CP01-200 | 50 ug x 4 tubes | Lyophilized | 4 tubes of 6 uM x 50 ul | |
| CP04-100 | 20 ug x 5 tubes | Liquid | ||
| High conc WT Cas9 (5 mg/ml after reconstitution) | CP02 | 250 ug (1500 pmol) | Lyophilized | 30 uM x 50 ul |
| CP03 | 250 ug x 4 tubes | Lyophilized | 4 tubes of 30 uM x 50 ul | |
| CP04-500 | 100 ug x 5 tubes | Liquid | ||
| D10A Nickase protein | CN01 | 50 ug (300 pmol) | Lyophilized | 6 uM x 50 ul |
| CN02 | 250 ug (1500 pmol) | Lyophilized | 30 uM x 50 ul | |
| D10A/H840 dCas9 protein | CD01 | 50 ug (300 pmol) | Lyophilized | 6 uM x 50 ul |
| CD02 | 50 ug x 5 tubes | Lyophilized | 5 tubes of 6 uM x 50 ul | |
| High fidelity SniperCas9 protein | CS01 | 50 ug (300 pmol) | Lyophilized | 6 uM x 50 ul |
| CS02 | 250 ug (1500 pmol) | Lyophilized | 30 uM x 50 ul | |
| Cy3 WT Cas9 protein | CP06 | 25 ug (150 pmol) | Liquid | |
| Cas9 reaction buffer 10x | CB01 | 500 ul | Liquid | |
| Cas9 dilution buffer | CB03 | 1 ml | Liquid |
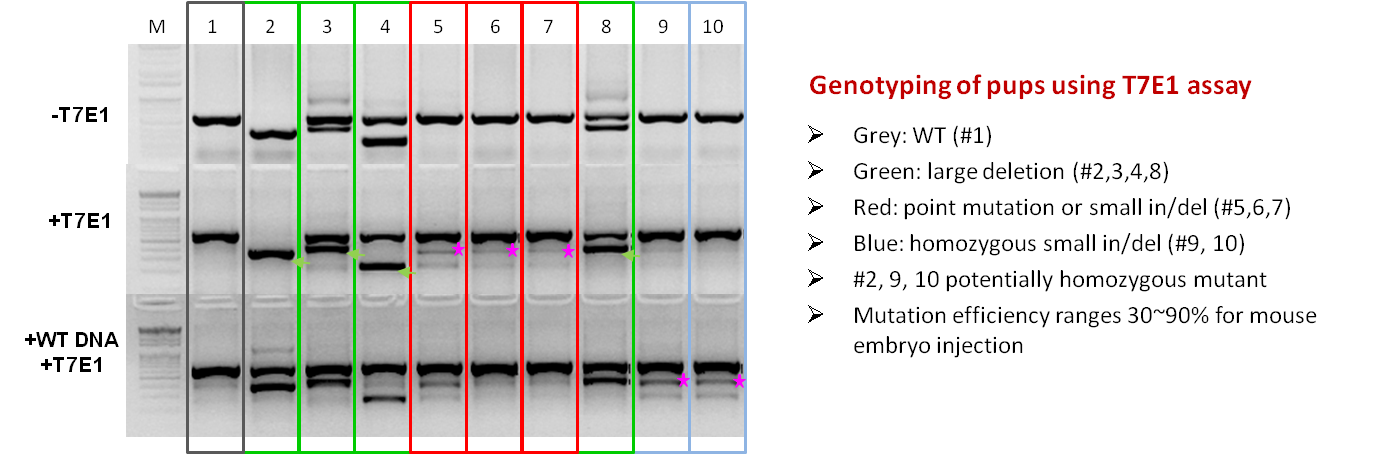
Injection of RNP complex
Publications referring our Cas9 protein
- Peptide Nucleic Acid-Mediated Regulation of CRISPR-Cas9 Specificity. Carufe KEW et al. (2024) Nucleic Acid Ther 34(5):245-256.
- DNMT1 mutant ants develop normally but have disrupted oogenesis. Ivasyk I et al. (2023) Nat Commun 14(1): 2201.
- Spinal cord repair is modulated by the neurogenic factor Hb-egf under direction of a regeneration-associated enhancer. Cigliola V et al. (2023) Nat Commun 14:4857.
- CRISPR/Cas9 mediated genome editing of Caenorhabditis nigoni using the conserved dpy-10 co-conversion marker. Choi CP & Villeneuve AM (2023) microPublication Biology 10:17912.
- CRISPR-enhanced human adipocyte browning as cell therapy for metabolic disease. Tsagkaraki E et al. (2021) Nat Commun 26; 12(1): 6931.
- Regulatory gene function handoff allows essential gene loss in mosquitoes. Cheatle AM et al. (2020) Commun Biol 3(1): 540.
- CRISPR/Cas9-mediated knock-in of alligator cathelicidin gene in a non-coding region of channel catfish genome. Simora RMC et al. (2020) Sci Rep 10(1): 22271.
- Simple embryo injection of long single-stranded donor templates with the CRISPR/Cas9 system leads to homology-directed repair in Xenopus tropicalis and Xenopus laevis. Nakayama T et al. (2020) Genesis 58(6): e23366.
- Multiplex CRISPR/Cas screen in regenerating haploid limbs of chimeric Axolotls. Sanor LD et al (2020) eLife 9: e48511.
- Germline CRISPR/Cas9-mediated gene editing prevents vision loss in a novel mouse model of aniridia. Mohanna SZM et al (2020) Mol Ther Methods Clin Dev 17: P478-490.
- Non-viral delivery of CRISPR/Cas9 complex using CRISPR-GPS nanocomplexes. Jain PK et al (2019) Nanoscale 11:21317-21323.
- Programmable CRISPR-Cas repression, activation, and computation with sequence-independent targets and triggers. Jin M et al. (2019) ACS Synthetic Biology 8(7):1583-1589.
- CRISPR/Cas9 gene editing in the West Nile Virus vector, Culex quinquefasciatus Say. Anderson ME at al. (2019) pLoS One 0224857.
- CRISPR/Cas9 Genome Editing Introduction and Optimization in the Non-model Insect Pyrrhocoris apterus. Kotwica-Rolinska J et al. (2019) Front Physiol 10: 00891.
- Small molecule agonists of Ae. aegypti neuropeptide Y receptor block mosquito biting. Duvall LB et al. (2019) Cell 176(4): 687.
- CRISPR/Cas9 F0 Screening of Congenital Heart Disease Genes in Xenopus tropicalis. Engin D et al. (2018) Methods in Molecular Biology 1865: 163.
- Microinjection of CRISPR/Cas9 Protein into Channel Catfish, Ictalurus punctatus, Embryos for Gene Editing. Elaswad A et al. (2018) J Vis Exp (131): e56275.
- Tissue-specific gene inactivation in xenopus laevis: knockout of lhx1 in the kidney with CRISPR/Cas9. DeLay BD et al. (2018) Genetics 208(2):673-686.
- A Non-integrating lentiviral approach overcomes cas9-induced immune rejection to establish an immunocompetent netastatic renal cancer model. Hu J et al. (2018) Mol Ther Methods Clin Dev 23(9):203-210.
- High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Wagner DL et al. (2018) Nature Medicine 25:242–248.
- Application and optimization of CRISPR–Cas9-mediated genome engineering in axolotl (Ambystoma mexicanum) Fei J et al. (2018) Nat Protocols 13: 2908–2943.
- One‐step CRISPR/Cas9 method for the rapid generation of human antibody heavy chain knock‐in mice. Lin YC et al. (2018) 37:e99243 EMBO J 37:e99243.
- CRISPR knockouts reveal an endogenous role for ancient neuropeptides in regulating developmental timing in a sea anemone. Nakanishi N & Martindale MQ (2018)eLife 7: e39742.
- Antagonistic BMP–cWNT signaling in the cnidarian Nematostella vectensis reveals insight into the evolution of mesoderm. Wijesena N et al. (2017) Proc Natl Acad Sci USA 114: E5608–E5615.
- Generation of heritable germline mutations in the jewel wasp Nasonia vitripennis using CRISPR/Cas9. Li M et al. (2017) Scientific Reports 7:901.
- Generation of biallelic F0 mutants in medaka using the CRISPR/Cas9 system. Sawamura R et al. (2017) Genes to Cells 22:756–763.
- Laminar flow downregulates Notch activity to promote lymphatic sprouting. Choi D et al. (2017) J Clin Invest 127(4):1225-1240.
- Localized TWIST1 and TWIST2 basic domain substitutions cause four distinct human diseases that can be modeled in C. elegans. Kim S et al. (2017) Hum Mol Genet 26(11): 2118–2132.
- Genetic Basis of Melanin Pigmentation in Butterfly Wings. Zhang L et al. (2017) Genetics205(4):1537-1550.
- Efficient CRISPR/Cas9-assisted gene targeting enables rapid and precise genetic manipulation of mammalian neural stem cells. Bressan RB et al. (2017) Development 144(4):635-648.
- CRISPR/Cas9-mediated gene editing in human zygotes using Cas9 protein. Tang L et al. (2017) Mol Genetics Genomics 292(3):525-533.
- A truncating mutation in CEP55 is the likely cause of MARCH, a novel syndrome affecting neuronal mitosis. Frosk P et al. (2017) J Med Genet 54(7):490-501.
- Quantitative Analysis of Synthetic Cell Lineage Tracing Using Nuclease Barcoding. Schmidt ST et al. (2017) ACS Synth Biol 6(6):936–942.
- Targeted gene knock-in by CRISPR/Cas ribonucleoproteins in porcine zygotes. Park KE et al. (2017) Hum Mol Genet 7:42458.
- Maternal Supply of Cas9 to Zygotes Facilitates the Efficient Generation of Site-Specific Mutant Mouse Models. Cebrian-Serrano A et al. (2017) PLoS One0169887.
- Molecular logic behind the three-way stochastic choices that expand butterfly colour vision. Perry M et al. (2016) Nature 535(7611):280-4.
- Leapfrogging: primordial germ cell transplantation permits recovery of CRISPR/Cas9-induced mutations in essential genes. Blitz IL et al. (2016) Development 143(15):2868-75.
- Delivery of Cas9 Protein into Mouse Zygotes through a Series of Electroporation Dramatically Increases the Efficiency of Model Creation. Wang W et al. (2016) J Genetics Genomics 43(5)319-327.
- Efficient genome engineering approaches for the short-lived African turquoise killifish. Harel I et al. (2016) Nature Protocols. 11(10):2010-28.
- Rapid Screening for CRISPR-Directed Editing of the Drosophila Genome Using white Co-Conversion. Ge DT et al. (2016) G3 (Bethesda) 6(10): 3197–3206.
- A truncating mutation in CEP55 is the likely cause of MARCH, a novel syndrome affecting neuronal mitosis. Frosk P et al. (2016) J Medical Genetics 104296.
- Generation and characterization of tamoxifen-inducible Pax9-creER knock-in mice using CrispR/Cas9. Jifan F et al. (2016) Genesis 54: 490–496.
- Expanding the genetic toolkit in Xenopus: Approaches and opportunities for human disease modeling. Tandon P et al. (2016) Dev Biology S0012-1606.
- Dual daughter strand incision is processive and increases the efficiency of DNA mismatch repair. Hermans N et al. (2016) Nucleic Acids Res 44(14):6770-86.
- Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Zhang L & Reed RD (2016) Nat Commun 7:11769.
- Pre-bilaterian origin of the blastoporal axial organizer. Kraus Y et al. (2016) Nat Commun 7:11694.
- CRISPRs for optimal targeting: delivery of CRISPR components as DNA, RNA, and protein into cultured cells and single-cell embryos. Kouranova E et al. (2016) Hum Gene Ther 27(6):464-75.
- Highly efficient genome editing of murine and human hematopoietic progenitor cells by CRISPR/Cas9. Gundry MC et al. (2016) Cell Reports 17(5):1453-61.
- Genomic Access to Monarch Migration Using TALEN and CRISPR/Cas9-Mediated Targeted Mutagenesis. Markert MJ et al. (2016)G3 (Bethesda) 6(4):905-15.
- A CRISPR Path to Engineering New Genetic Mouse Models for Cardiovascular Research. Miano JM et al. (2016) Arterioscler Thromb Vasc Biol 36(6):1058-75.
- CRISPR/Cas9-mediated mutagenesis in the sea lamprey Petromyzon marinus: a powerful tool for understanding ancestral gene functions in vertebrates. Square T et al. (2015) Development 142(23):4180-7.
- High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR/Cas9 Ribonucleoprotein Complexes. Paix A et al. (2015) Genetics 201(1):47-54.
- CRISPR-CAS9 D10A nickase target-specific fluorescent labeling of double strand DNA for whole genome mapping and structural variation analysis. McCaffrey J et al. (2015) Nucleic Acids Res 44(2):e11.
- Cell lineage tracing using nuclease barcoding. Schmidt ST et al. (2016) Cornell University Library Epub.
- Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Basu S et al. (2015) Proc Natl Acad Sci U S A 112(13):4038-43.
- In vivo Modeling Implicates APOL1 in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress. Anderson BR et al. (2015) PLoS Genet 11(7):e1005349.
- CRISPR/Cas9: An inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Bhattacharya D et al. (2015) Dev Biology 408(2): 196-204.
- Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Kistler KE et al. (2015) Cell Reports 11(1): 51-60.
- Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses. Dan W et al. (2015) Hum Gene Ther 26(7): 432-442.
- Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Hendel A et al. (2015) Nat Biotech 33:985-89.
- Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Aida T et al. (2015) Genome Biology 16:87.
- TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Ikmi A et al. (2014) Nat Commun 24(5):5486.
- Chapter Seventeen: Cas9-Based Genome Editing in Xenopus tropicalis. Nakayama T et al. (2014) Methods Enzymol (Editor J A Doudna & E. J. Sontheimer) 546:355-375.
Books referring our Cas9 protein
- Genome Editing & Engineering: From TALENs, ZFNs and CRISPRs to Molecular Surgery (ed. Appasani K) Leapfrogging: Targeting CRISPR/Cas9 Genome Editing to the Germline in Xenopus. Blitz IL (2018) Cambridge University Press
- Diversity and Evolution of Butterfly Wing Patterns (ed. Sekimura T & Nijhout HF) A Practical Guide to CRISPR/Cas9 Genome Editing in Lepidoptera. Zhang L & Reed RD (2017) pp 155-172. Springer
- The Cricket as a Model Organism; Development, Regeneration, and Behavior by Horch, H.W., Mito, T., Popadić, A., Ohuchi, H., Noji, S. (2017) Springer
- Methods in Cell Biology, The Zebrafish: Genetics, Genomics, and Transcriptomics, Volume 135, 4th Edition (Detrich HW, Zon L. & Westerfield M) (2016) Academic Press
On-line manuals using our Cas9 protein
- Practical guide to genome-engineering with CRISPR-Cas9 in the mosquito Aedes aegypti (Kistler KE et al.) – Rockefeller University
- KI mice generation – Beth Israel Deaconess Medical Center Transgenic Core Facility
- RNA guide prep and purification – University of Wisconsin-Madison Biotechnology Center Transgenic Animal Facility
- Optimization of DNA, RNA and RNP Delivery for Efficient Mammalian Cell Engineering – Mirus Bio
- CRISPR-based gene disruptionin human hematopoietic cells – The Goodell Laboratory
| Cat No | Description | Size | Price | Order |
| CP01-50 | WT Cas9 Protein | 50 ug (300 pmol) | $ 95.00 | Add to cart |
| CP01-200 | WT Cas9 Protein | 50 ug x 4 tubes | $ 325.00 | Add to cart |
| CP02 | WT Cas9, High Conc | 250 ug (1500 pmol) | $ 340.00 | Add to cart |
| CP03 | WT Cas9, High Conc | 250 ug x 4 tubes | $ 1150.00 | Add to cart |
| CP04-100 | WT Cas9 Protein, Liquid Form | 20 ug x 5 tubes | $ 220.00 | Add to cart |
| CP04-500 | WT Cas9, High Conc, Liquid Form | 100 ug x 5 tubes | $ 695.00 | Add to cart |
| CN01 | Nickase Cas9 Protein | 50 ug (300 pmol) | $ 115.00 | Add to cart |
| CN02 | Nickase Cas9 Protein, High Conc | 250 ug (1500 pmol) | $ 350.00 | Add to cart |
| CD01 | dCas9 Protein | 50 ug (300 pmol) | $ 115.00 | Add to cart |
| CD02 | dCas9 Protein | 50 ug x 5 tubes | $ 350.00 | Add to cart |
| CS01 | High Fidelity Sniper Cas9 Protein | 50 ug (300 pmol) | $ 120.00 | Add to cart |
| CS02 | High Fidelity Sniper Cas9, High Conc | 250 ug (1500 pmol) | $ 385.00 | Add to cart |
| CB01 | 10x Cas9 Reaction Buffer | 500 ul | $ 28.00 | Add to cart |
| CB03 | Cas9 Dilution Buffer | 1 ml | $ 26.00 | Add to cart |