Mutyper
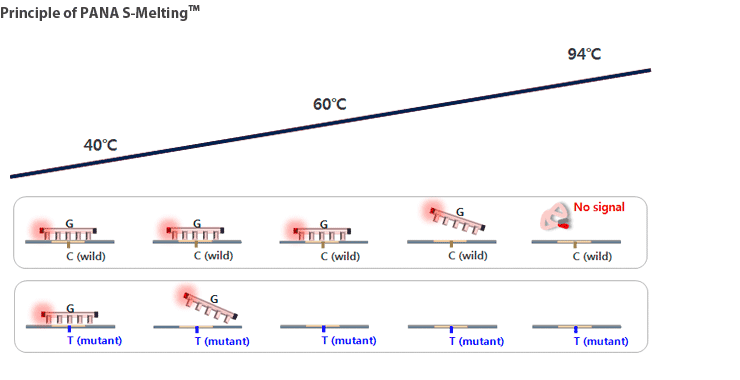
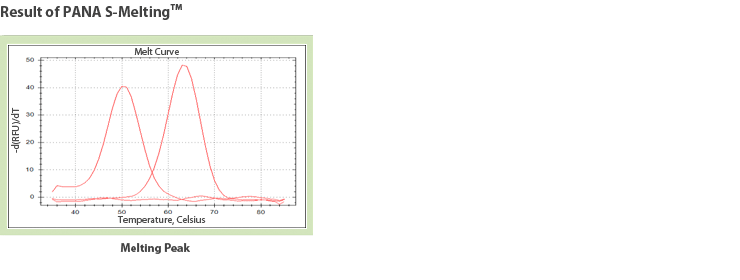
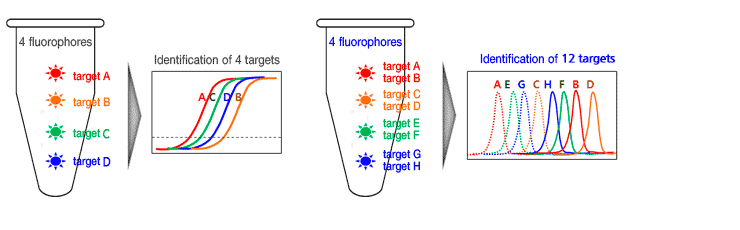
Therefore, PANAMutyper™ R technology can detect a single base mutation accurately with high sensitivity for only a minute amount of mutant type DNA, such as the samples from blood. It is also very fast and convenient method for detecting mutant.
Lung cancer is a metastatic cancer which the cancer cells are developed in the bronchial tubes or alveoli and grow along the blood vessel or lymphatic vessel. There are two types of lung cancer based on the sizes and shape of the cancer cell; non-small cell lung cancer (NSCLC) takes 75% and small cell lung cancer takes 25%. Usually, it is hard to find lung cancer in the early stage and is diagnosed as lung cancer when it is already progressed. So the prognosis tends to be bad. It is known that the patients with EGFR mutation show very high drug responses against EGFR tyrosine kinase inhibitor (TKI) such as Gefitinib (Iressa, AstraZenca) and Erlotinib (Tarceba, Roche). We expect that genotyping EGFR genes of the lung cancer patients enables predicting drug response before treatment and this will lead effective lung cancer treatment.
|
PANAMutyper™ R EGFR |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
EGFR Mutation Details |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
KRAS mutation is found in several cancers such as pancreatic cancer, colorectal cancer, lung cancer, biliary tract cancer and thyroid cancer. The existence of KRAS mutations is often related with a prognostic marker to drug response. For example, KRAS mutation is considered a strong prognostic marker for drug response of tyrosine kinase inhibitors such as Gefitinib (Iressa) or Erlotinib (Tarceva).Recently, KRAS mutation is often detected in colorectal cancer and may be related with drug responseto Cetuximab (Erbitux) or Panitumumab (Vectibix) that is used for colon cancer therapy. Therefore,examination of KRAS mutation is needed to determine drug resistance of patients with colorectal or lung cancers and will be helpful for cancer therapies.
|
PANAMutyper™ R KRAS |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
KRAS Mutation Details |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
NRAS mutation is found in several cancers such as melanoma (13~25%), colorectal cancer (1~6%), lung cancer (1%), thyroid cancer (7%) and hepatocellular carcinoma (10%). It is know that the drug response against colorectal cancer medicine such as Erbitux and Cetuximab decreased and the prognosis of the metastatic colorectal cancer patient is bad if the patient has NRAS mutation. Recently, there are some papers that report the drug responses can be different for the NRAS mutation type so it is important to genotype each mutation. NRAS gene mutation genotyping test is necessary especially for predicting the drug sensitivity and prognosis. This test expected to play a major role for predicting the treatment of the patients and prognosis.
|
PANAMutyper™ R NRAS |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
NRAS Mutation Details |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Please email the quote request to order@pnabio.com.